BiG-SCAPE / CORASON tutorial
This tutorial explains how to download datasets to run BiG-SCAPE and CORASON, and provides examples for how to use both tools.
Example dataset
The example dataset consists on three parts: a set of genomes (includes a single cluster from MIBiG), a set of GenBank files (selected clusters predicted from the set of genomes) and the TauD sequence from a Streptomyces genome (NRRL B-1347).
Download and execute the following script to obtain the files:
$ mkdir ~/bin/example # not required if you already have that
$ curl https://raw.githubusercontent.com/nselem/master/scripts/data_bigscape_corason.sh
$ mv data_bigscape_corason.sh ~/bin/example/data_bigscape_corason.sh
$ chmod a+x ~/bin/example/data_bigscape_corason.sh
$ cd ~/bin/example && ~/bin/example/data_bigscape_corason.sh
The content of the ~/bin/example folder should look like this:

Data can also be downloaded manually at:
How to compile your own input dataset
In this tutorial, we will work with the above example data. If you want to use your own gene clusters as input for BiG-SCAPE and the CORASON family mode integrated within the BiG-SCAPE pipeline, you can search for publicly available genomes in
antiSMASH-DB and download the desired cluster files in GenBank (.gbk) format. Alternatively, you can perform your own antiSMASH runs on the public web server or on your local system, collect the cluster GBK files and put them together in a folder that you can use as input for BiG-SCAPE. Entries from MIBiG can be added automatically by adding the --mibig flag to the end of your BiG-SCAPE command (see below).
BiG-SCAPE v1 example
For the most up-to-date tutorial on how to use BiG-SCAPE, please see the Wiki section of the official repository site.
We will now proceed with the example data. Once data has been downloaded, run the following command at the terminal (you should be now in the ~/bin/example folder):
$ run_bigscape gbks example_output
If everything goes well, the terminal will show something similar to:
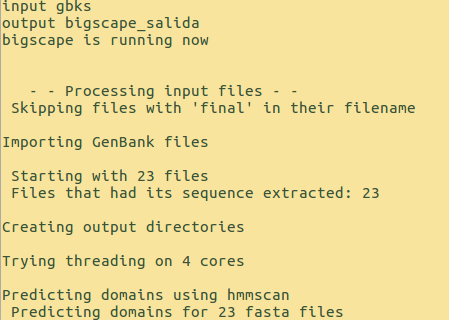
The input for BiG-SCAPE is the directory gbks that contains GenBank files of sequences of Biosynthetical gene clusters (BGCs) predicted by antiSMASH. The BiG-SCAPE output will be stored in the directory example_output.
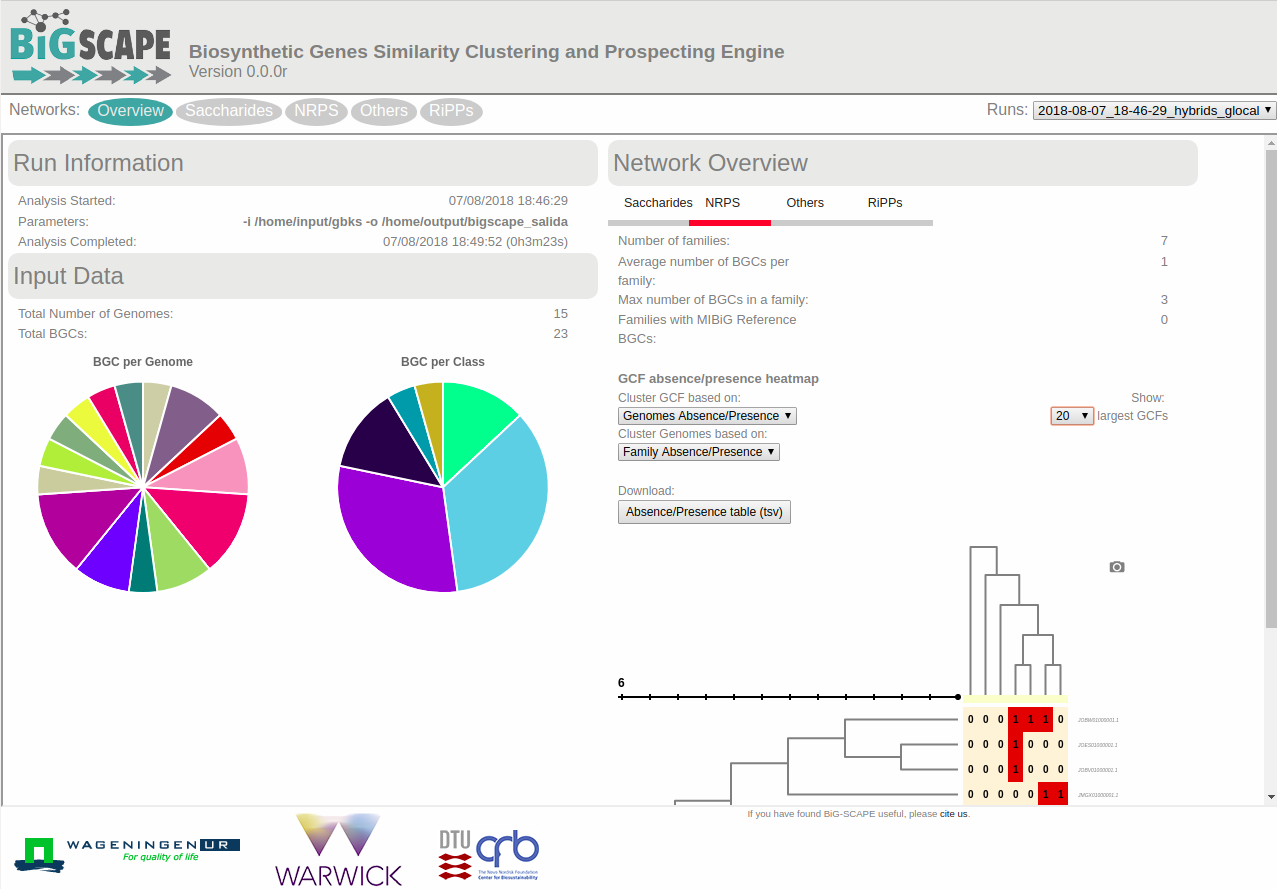
After BiG-SCAPE has finished successfully open the index.html file located inside the example_output folder with your browser, (e.g. Chrome or Firefox). The file contains an interactive offline webpage that displays the BiG-SCAPE results and allows you to explore them.
To start exploring, select a class at the top of the site:
Now, the screen will display a network visualization of BGC families within this class.
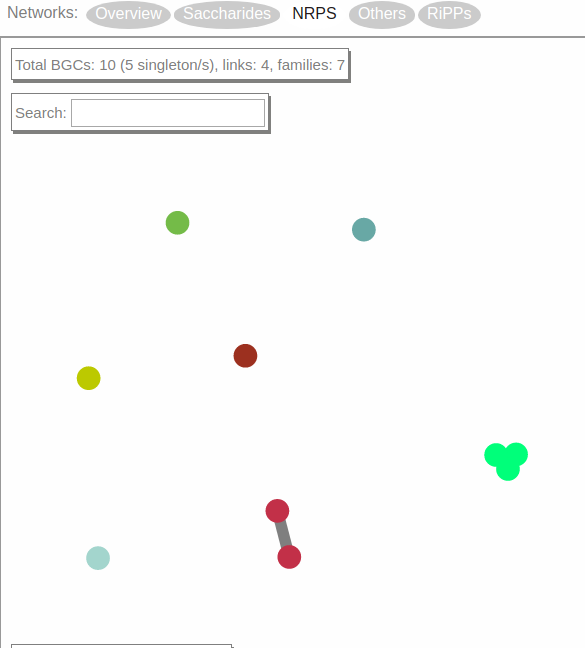
In this case, the NRPS class contains 10 BGCs organized in one gene cluster family of three members, one family of two members and five singletons.
Now select a family in this network to visualize BGCs sorted and aligned CORASON-like.
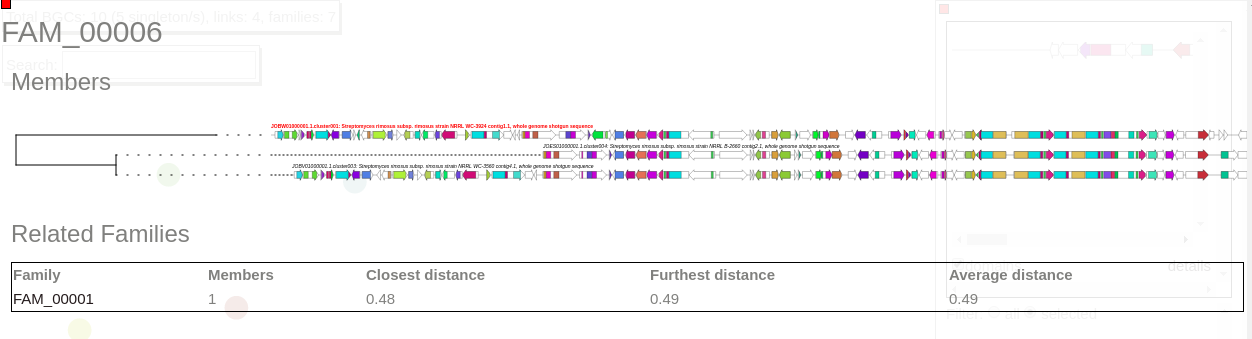
This family contains three members.
BiG-SCAPE output (.network files) can also be imported with Cytoscape.
In another example BiG-SCAPE was employed to calculate BGC familiess in 103 complete Streptomyces genomes. The outcome of this run can be found here
CORASON example
$ ~/bin/run_corason -q queryFile gbksDirectory referenceBGC -g
After getting the example data, the next step to identify variations in the genomic vicinity of tauD that remain similar to the reference BGC JMGX01000001.1.cluster003.gbk is to execute CORASON:
$ ~/bin/run_corason TauD.fasta gbks gbks/JMGX01000001.1.cluster003.gbk
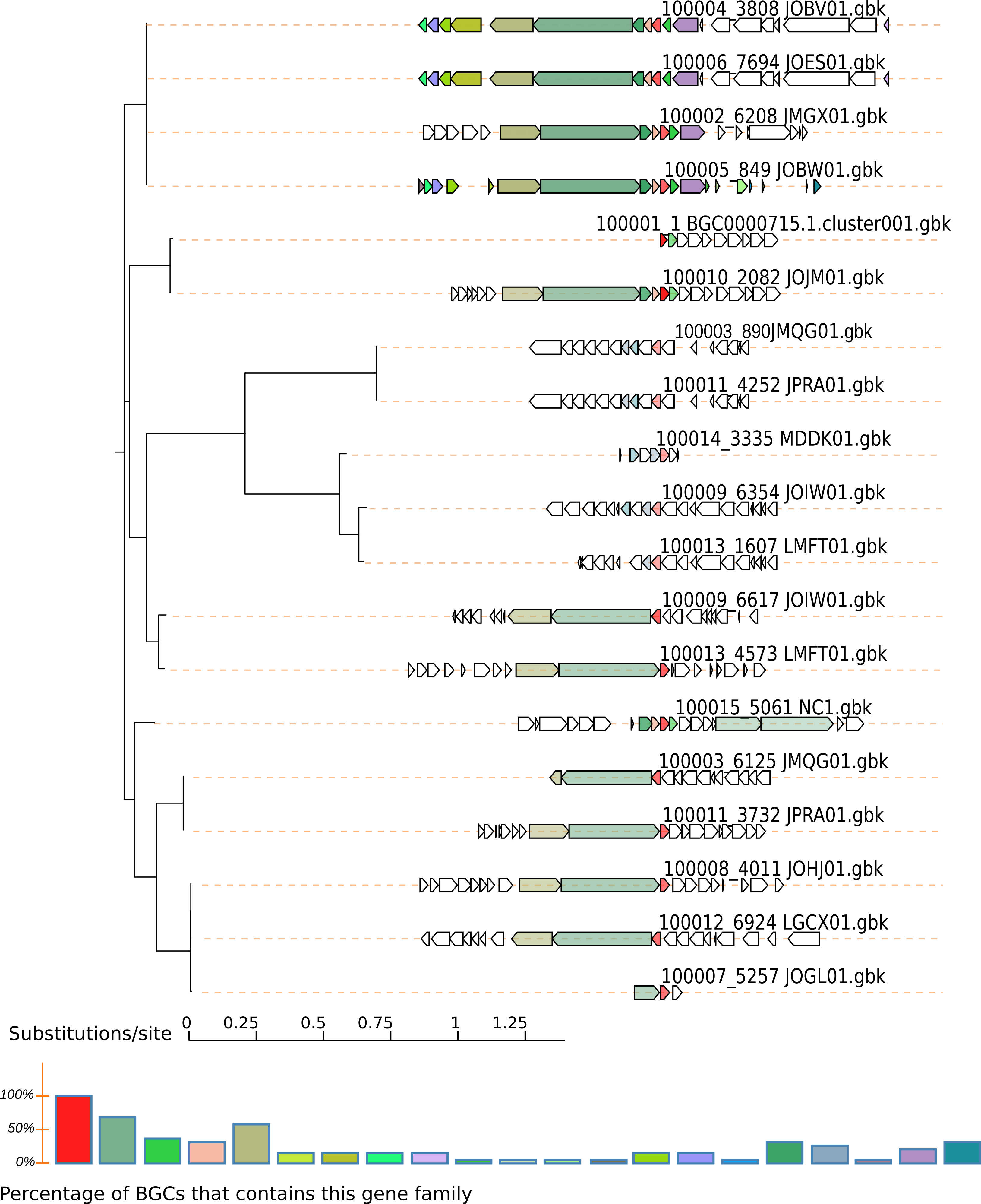
The output will be in the directory query-output, in this case TauD.fasta-output. See the svg output (e.g. $ firefox TauD.fasta-output/Joined.svg). In this case we can see even fragment BGCs without the NRPS genes:
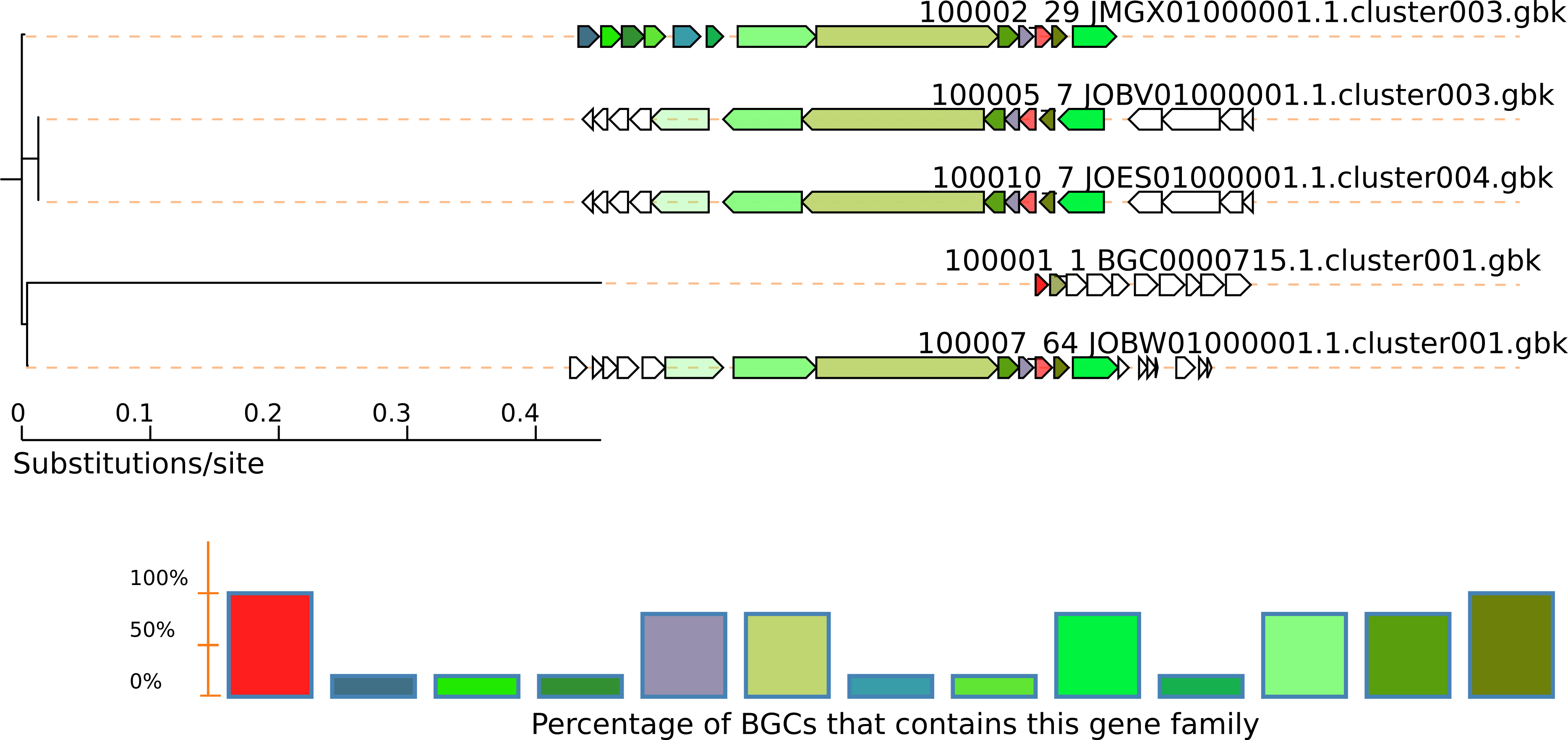
To obtain more variations of genomic vicinities, switch the exploration to genomes instead of BGCs. Here, more BGCs are found and differences are not limited to gene content of the BGC; differences in sequence similarity are also indicated by a color gradient.
~/bin/run_corason TauD.fasta genomes genomes/JOBW01.gbk