Installation
BiG-SCAPE can be installed manually (i.e. by installing dependencies and running the code directly) or by using Docker. CORASON can be run using Docker.
Docker
Docker is a container platform provider available for multiple operating systems. If you already have Docker engine installed, or plan to use another method to install the software, please skip this step.
We recommend checking the latest instructions for installing Docker in the official website.
This is a Linux minimal docker installation guide which uses curl (See curl installation if you do not have it installed):
$ curl -fsSL https://get.docker.com/ | sh
Then type (without brackets):
$ sudo usermod -aG docker [your username]
Important step: Log out from your session (restart your machine) and get back in into your user session before the next step. You may need to restart your computer and not just log out from your session in order to changes to take effect. Test your docker engine with the command:
$ docker run hello-world
BiG-SCAPE installation
For the most up-to-date instructions to install BiG-SCAPE, please see the Wiki section of the official repository site.
CORASON installation
Docker installation
$ mkdir ~/bin # not required if you already have that
$ curl -q https://raw.githubusercontent.com/nselem/corason/master/run_corason > ~/bin/run_corason
$ chmod a+x ~/bin/run_corason
$ ~/bin/run_corason
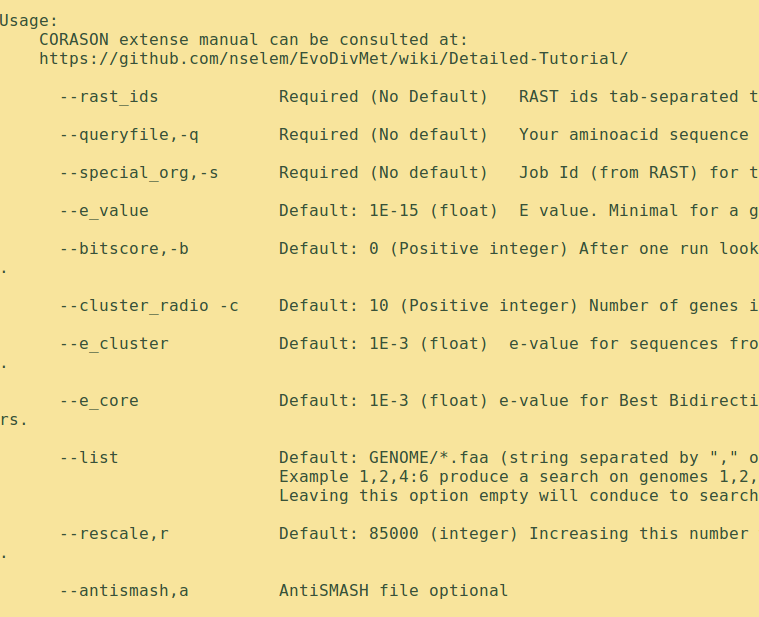
If CORASON is correctly installed the terminal displays the help menu after some seconds: